Outline of the post:
- What is Whitening or Sphering? Why?
- Steps to Whiten a dataset
- Mathematical intuition
- Implementation of Whitening in Python
What is Whitening or Sphering? Why?
“A whitening transformation or sphering transformation is a linear transformation that transforms a vector of random variables with a known covariance matrix into a set of new variables whose covariance is the identity matrix, meaning that they are uncorrelated and each have variance.The transformation is called “whitening” because it changes the input vector into a white noise vector.”
– Whitening Transformation, Wikipedia, 2020
Whitening or Sphering is a data pre-processing step. It can be used to remove correlation or dependencies between features in a dataset. This may help to better train a machine learning model. Figure 1 below shows the effect of decorrelation and whitening on a data that has correlated features (‘orange’ color dots).
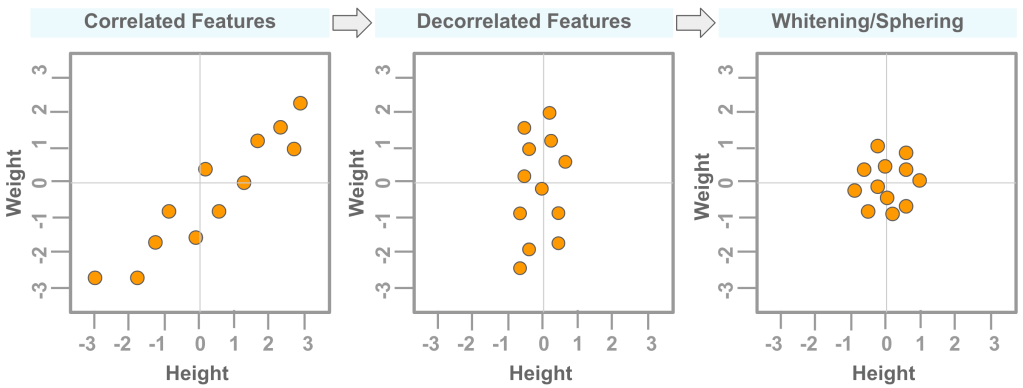
Steps to Whiten a dataset
Eigenvalues and eigenvectors are first calculated from the covariance of a zero centered data set. These are then used for Whitening the data using either PCA (principal component analysis) or ZCA (zero component analysis method).

- Step # 1: Find if data has one feature per row or one feature per column
- Step # 2: Zero-center the dataset
- Step # 3: Calculate the Covariance matrix using the zero-centered dataset
- Step # 4: Calculate the Eigenvalues and Eigenvectors
- Step # 5: Apply the Eigenvalues and Eigenvectors to the data for whitening transform.
Mathematical Intuition
Zero-center data:
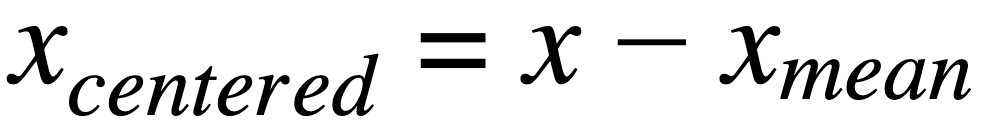
where,
xcentered: centered data for a featurex: is individual data point for a featurexmean: mean value for a feature
Covariance matrix:
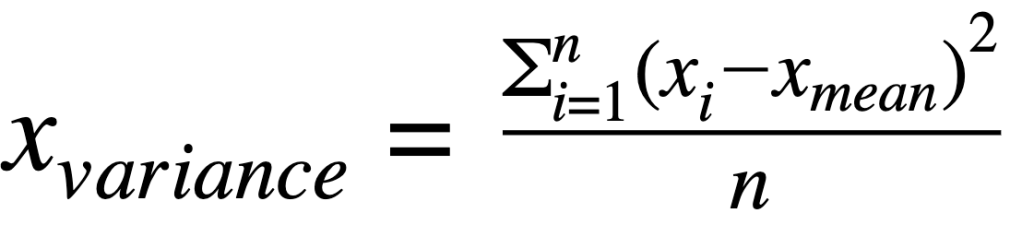
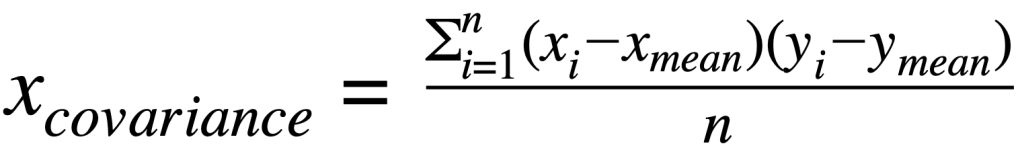
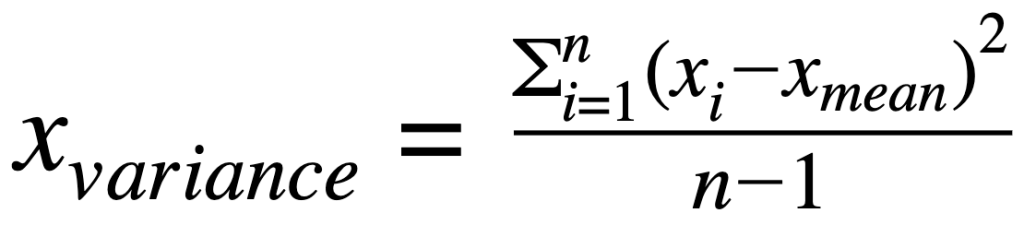
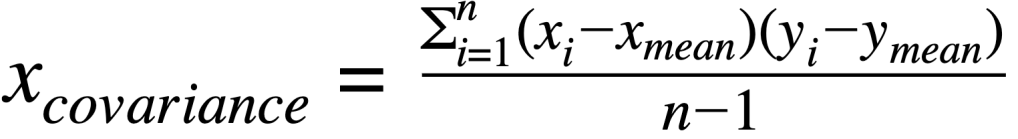
where:
xvariance: is variance for a featurexcovariance: is covariance between featuresxandyxiandyi: is individual data point for featurexandyxmeanand ymean: mean value for featuresxandy∑: indicates sum of valuesn: is the number of observations for a feature
Eigenvalues and Eigenvectors:

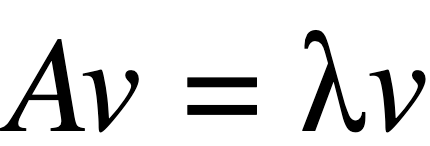
where:
A: Covariance matrix of featuresλ: is the matrix for EigenvaluesI: is identity matrixv: is the matrix for Eigenvectors
Whitening using PCA:
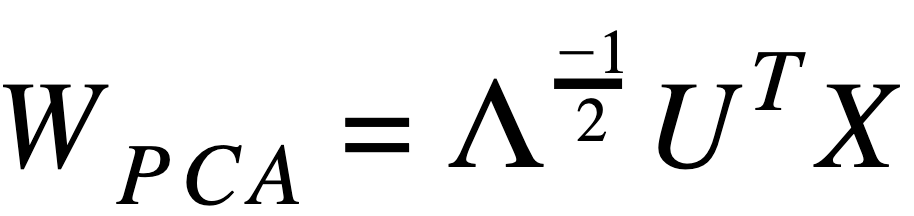
where:
X: Feature data- Λ: is the matrix for Eigenvalues (above is inverse square root)
- UT: is transpose of matrix for Eigenvectors
Whitening using ZCA:

where:
X: Feature data- Λ: is the matrix for Eigenvalues (above is inverse square root)
- UT: is transpose of matrix for Eigenvectors
Implementation of Whitening in Python
Example # 1: (with 5 data points)
# Import libraries
import numpy as np
import matplotlib.pyplot as plt
from scipy import linalg
# Create data
x = np.array([[1,2,3,4,5], # Feature-1: Height
[11,12,13,14,15]]) # Feature-2: Weight
print('x.shape:', x.shape)
# Center data
# By subtracting mean for each feature
xc = x.T - np.mean(x.T, axis=0)
xc = xc.T
print('xc.shape:', xc.shape, '\n')
# Calculate covariance matrix
xcov = np.cov(xc, rowvar=True, bias=True)
print('Covariance matrix: \n', xcov, '\n')
# Calculate Eigenvalues and Eigenvectors
w, v = linalg.eig(xcov)
# Note: Use w.real.round(4) to (1) remove 'j' notation to real, (2) round to '4' significant digits
print("Eigenvalues:\n", w.real.round(4), '\n')
print("Eigenvectors:\n", v, '\n')
# Calculate inverse square root of Eigenvalues
# Optional: Add '.1e5' to avoid division errors if needed
# Create a diagonal matrix
diagw = np.diag(1/(w**0.5)) # or np.diag(1/((w+.1e-5)**0.5))
diagw = diagw.real.round(4) #convert to real and round off
print("Diagonal matrix for inverse square root of Eigenvalues:\n", diagw, '\n')
# Calculate Rotation (optional)
# Note: To see how data can be rotated
xrot = np.dot(v, xc)
# Whitening transform using PCA (Principal Component Analysis)
wpca = np.dot(np.dot(diagw, v.T), xc)
# Whitening transform using ZCA (Zero Component Analysis)
wzca = np.dot(np.dot(np.dot(v, diagw), v.T), xc)
# Plot zero-centered, rotated and whitened data
plt.scatter(xc[0,:], xc[1,:], s=150, label='centered', alpha=0.5)
plt.scatter(xrot[0,:], xrot[1,:], s=150, label='rotated')
plt.scatter(wpca[0,:], wpca[1,:], s=150, marker='+', label='wpca')
plt.scatter(wzca[0,:], wzca[1,:], s=150, marker='x', label='wzca')
plt.xlabel('Height', fontsize=16)
plt.ylabel('Weight', fontsize=16)
plt.rc('xtick',labelsize=16)
plt.rc('ytick',labelsize=16)
plt.legend()
plt.tight_layout()
plt.savefig('whiten_1.png', dpi=300)
x.shape: (2, 5)
xc.shape: (2, 5)
Covariance matrix:
[[2. 2.]
[2. 2.]]
Eigenvalues:
[4. 0.]
Eigenvectors:
[[ 0.70710678 -0.70710678]
[ 0.70710678 0.70710678]]
Diagonal matrix for inverse square root of Eigenvalues:
[[5.00000000e-01 0.00000000e+00]
[0.00000000e+00 4.74531328e+07]]

Example # 2: (1000 data points)
# Import libraries
import numpy as np
import matplotlib.pyplot as plt
from scipy import linalg
# Create data
np.random.seed(1)
mu = [0,0]
sigma = [[6,5], [5,6]]
n = 1000
x = np.random.multivariate_normal(mu, sigma, size=n)
print('x.shape:', x.shape, '\n')
# Zero center data
xc = x - np.mean(x, axis=0)
print(xc.shape)
xc = xc.T
print('xc.shape:', xc.shape, '\n')
# Calculate Covariance matrix
# Note: 'rowvar=True' because each row is considered as a feature
# Note: 'bias=True' to divide the sum of squared variances by 'n' instead of 'n-1'
xcov = np.cov(xc, rowvar=True, bias=True)
print
# Calculate Eigenvalues and Eigenvectors
w, v = linalg.eig(xcov) # .eigh()
# Note: Use w.real.round(4) to (1) remove 'j' notation to real, (2) round to '4' significant digits
print("Eigenvalues:\n", w.real.round(4), '\n')
print("Eigenvectors:\n", v, '\n')
# Calculate inverse square root of Eigenvalues
# Optional: Add '.1e5' to avoid division errors if needed
# Create a diagonal matrix
diagw = np.diag(1/(w**0.5)) # or np.diag(1/((w+.1e-5)**0.5))
diagw = diagw.real.round(4) #convert to real and round off
print("Diagonal matrix for inverse square root of Eigenvalues:\n", diagw, '\n')
# Calculate Rotation (optional)
# Note: To see how data can be rotated
xrot = np.dot(v, xc)
# Whitening transform using PCA (Principal Component Analysis)
wpca = np.dot(np.dot(diagw, v.T), xc)
# Whitening transform using ZCA (Zero Component Analysis)
wzca = np.dot(np.dot(np.dot(v, diagw), v.T), xc)
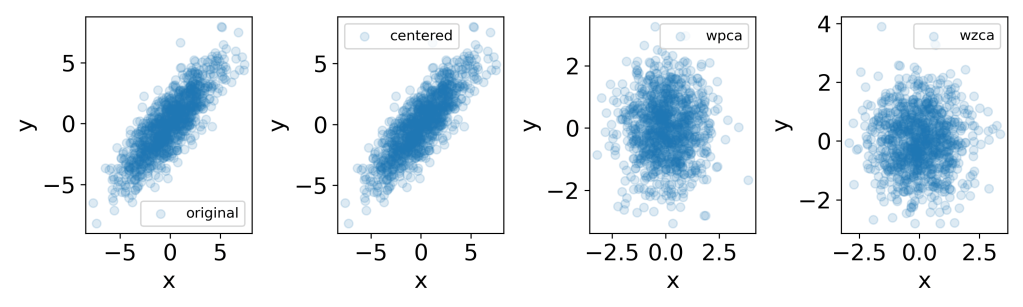
fig = plt.figure(figsize=(10,3))
plt.subplot(1,4,1)
plt.scatter(x[:,0], x[:,1], label='original', alpha=0.15)
plt.xlabel('x', fontsize=16)
plt.ylabel('y', fontsize=16)
plt.legend()
plt.subplot(1,4,2)
plt.scatter(xc[0,:], xc[1,:], label='centered', alpha=0.15)
plt.xlabel('x', fontsize=16)
plt.ylabel('y', fontsize=16)
plt.legend()
plt.subplot(1,4,3)
plt.scatter(wpca[0,:], wpca[1,:], label='wpca', alpha=0.15)
plt.xlabel('x', fontsize=16)
plt.ylabel('y', fontsize=16)
plt.legend()
plt.subplot(1,4,4)
plt.scatter(wzca[0,:], wzca[1,:], label='wzca', alpha=0.15)
plt.xlabel('x', fontsize=16)
plt.ylabel('y', fontsize=16)
plt.legend()
plt.tight_layout()
plt.savefig('whiten_2.png', dpi=300)
x.shape: (1000, 2)
(1000, 2)
xc.shape: (2, 1000)
Eigenvalues:
[ 1.005 11.1862]
Eigenvectors:
[[-0.71582163 -0.69828318]
[ 0.69828318 -0.71582163]]
Diagonal matrix for inverse square root of Eigenvalues:
[[0.9975 0. ]
[0. 0.299 ]]
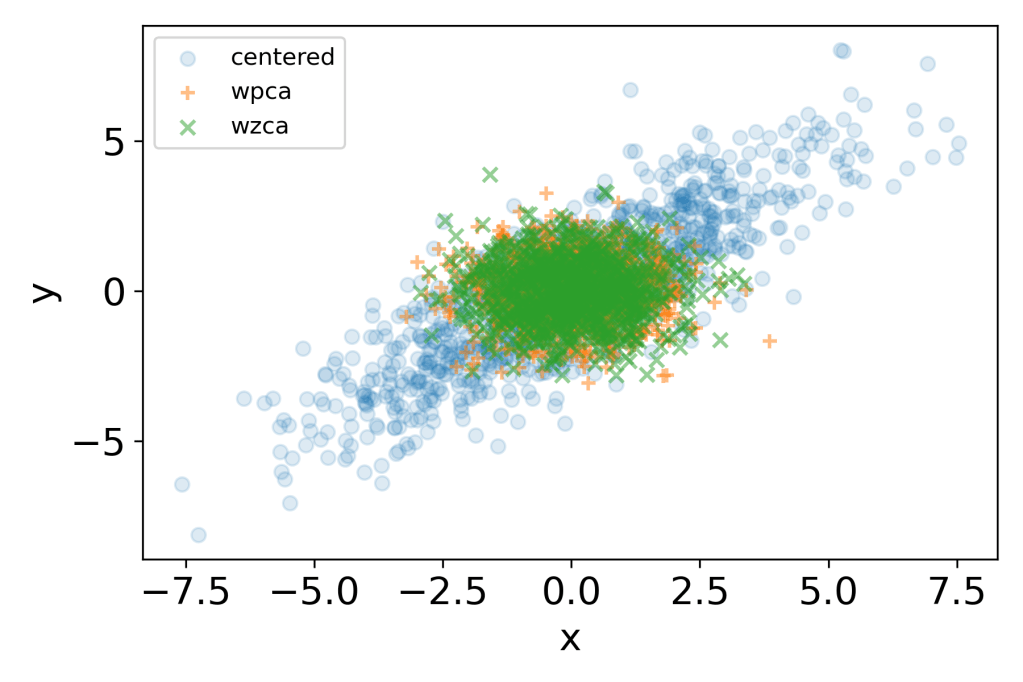
# Overlay plot
plt.scatter(xc[0,:], xc[1,:], label='centered', alpha=0.15)
plt.scatter(wpca[0,:], wpca[1,:], marker='+',label='wpca', alpha=0.5)
plt.scatter(wzca[0,:], wzca[1,:], marker='x', label='wzca', alpha=0.5)
plt.xlabel('x', fontsize=16)
plt.ylabel('y', fontsize=16)
plt.legend()
plt.tight_layout()
plt.savefig('whiten_3.png', dpi=300)
.
Check out the explanation on YouTube video!
